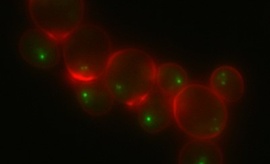
Spatial telomere organization and clustering in yeast Saccharomyces cerevisiae nucleus is generated by a random dynamics of aggregation–dissociation
Spatial and temporal behavior of chromosomes and their regulatory proteins is a key control mechanism in genomic function. This is exemplified by the clustering of the 32 budding yeast telomeres that form foci in which silencing factors concentrate. To uncover the determinants of telomere distribution, we compare live-cell imaging with a stochastic model of telomere dynamics that we developed. We show that random encounters alone are inadequate to produce the clustering observed in vivo. In contrast, telomere dynamics observed in vivo in both haploid and diploid cells follows a process of dissociation–aggregation. We determine the time that two telomeres spend in the same cluster for the telomere distribution observed in cells expressing different levels of the silencing factor Sir3 protein, limiting for telomere clustering. We conclude that telomere clusters, their dynamics, and their nuclear distribution result from random motion, aggregation, and dissociation of telomeric regions, specifically determined by the amount of Sir3.